Creation of a Staged Tree#
EventTree Class#
The first starting point in constructing a Chain Event Graph (CEG) is to create an event tree describing the process being studied. An event tree is a directed tree graph with a single root node. The nodes with no emanating edges are called leaves, and the non-leaf nodes are called situations.
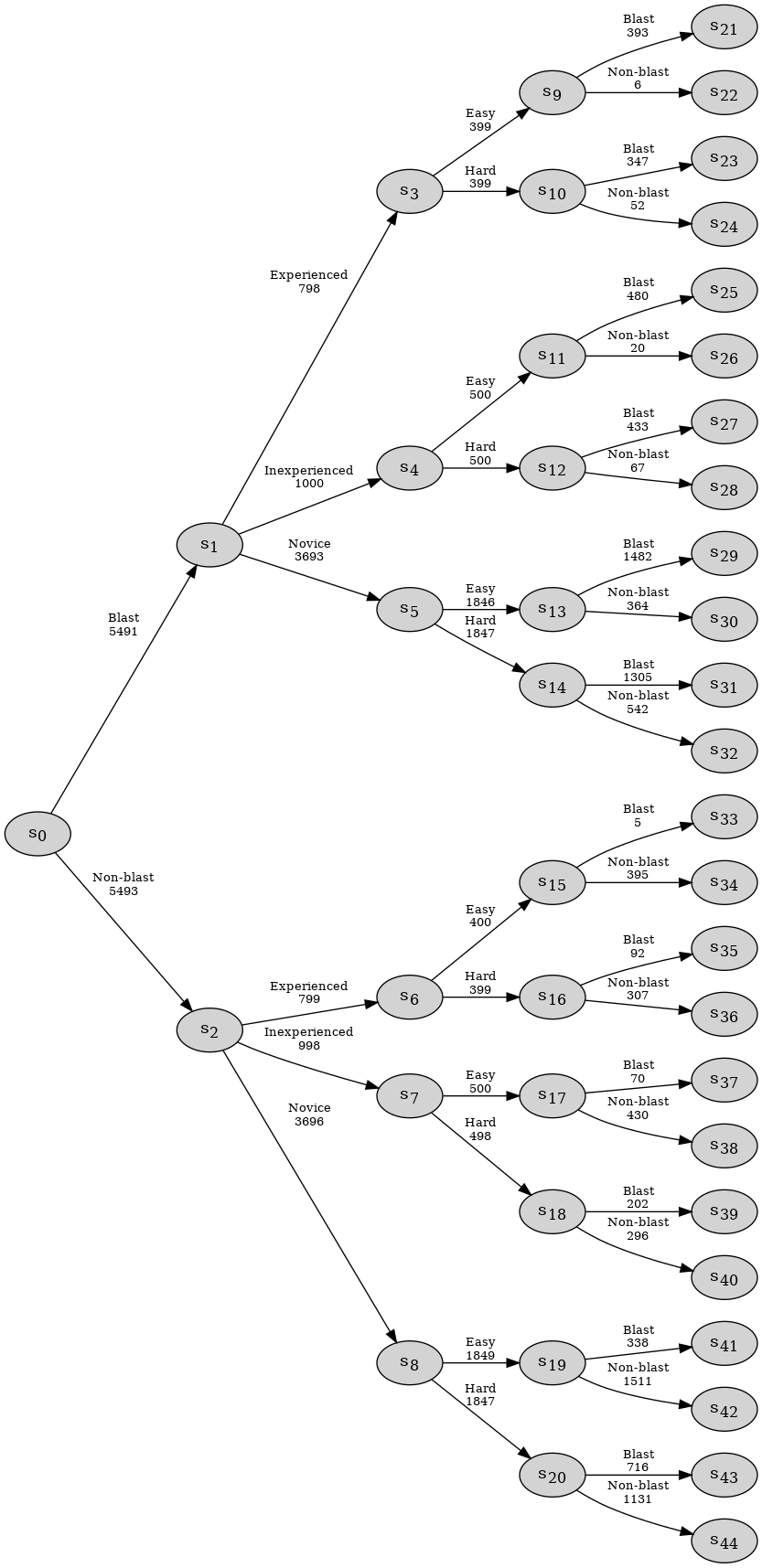
In this example we work with a data set which contains 4 categorical variables; Classification, Group, Difficulty, and Response.
Each individual is given a binary classification; Blast or Non-blast. Each group is rated on their experience level: Experienced, Inexperienced, or Novice. The classification task they are given has a difficulty rating of Easy or Hard. Finally, their response is shown: Blast or Non-blast.
We begin by importing the data set and initializing the EventTree object, as shown below:
from cegpy import EventTree
import pandas as pd
df = pd.read_excel('../../data/medical_dm_modified.xlsx')
print(df.head())
#initialize the event tree
et = EventTree(df)
Classification Group Difficulty Response
0 Blast Experienced Easy Blast
1 Non-blast Experienced Easy Non-blast
2 Non-blast Experienced Hard Blast
3 Non-blast Experienced Hard Non-blast
4 Blast Experienced Easy Blast
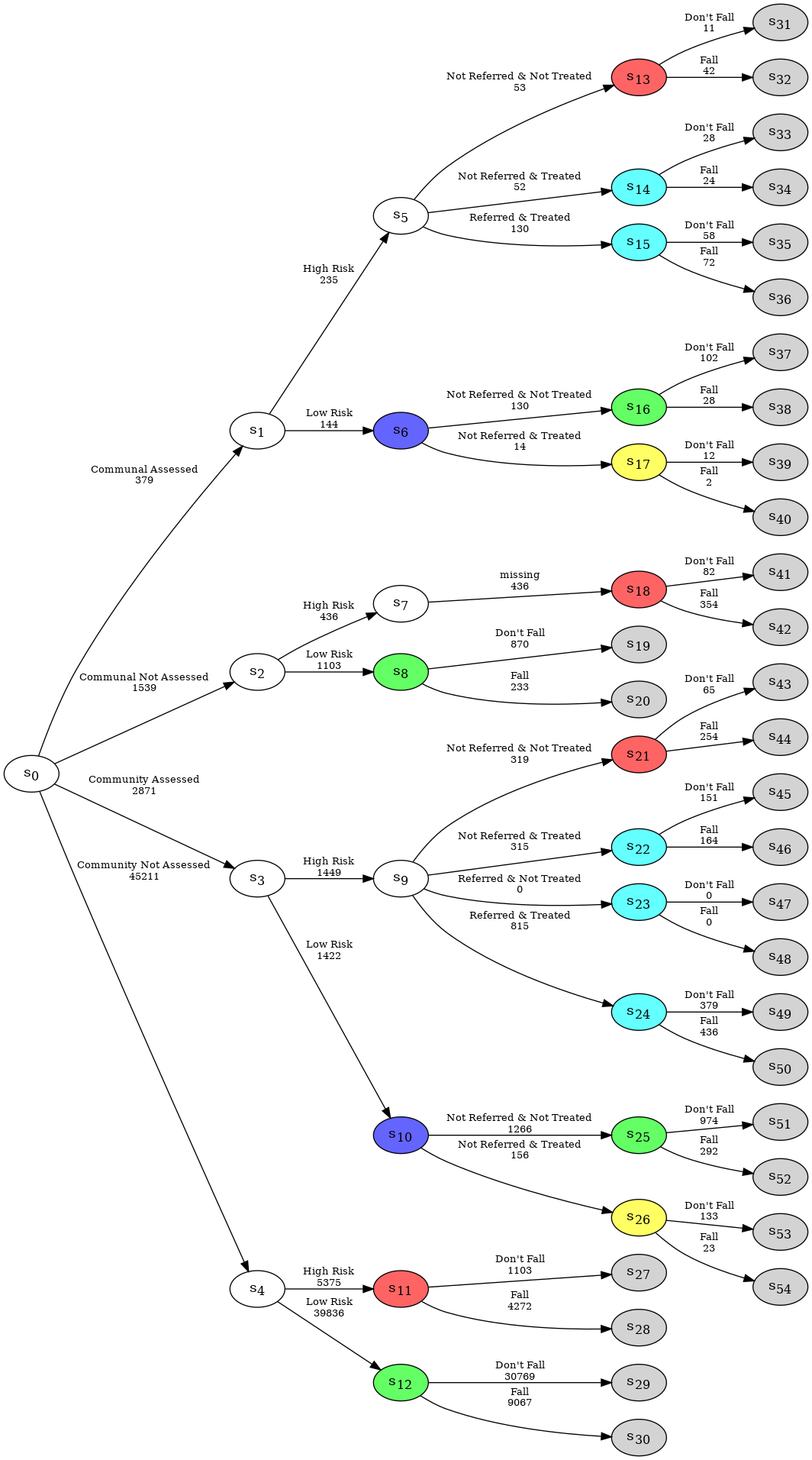
In order to display the EventTree, we can use the method create_figure(). The numbers above the edges of the event tree represent the number of individuals who passed through the given edge.
et.create_figure()
StagedTree Class#
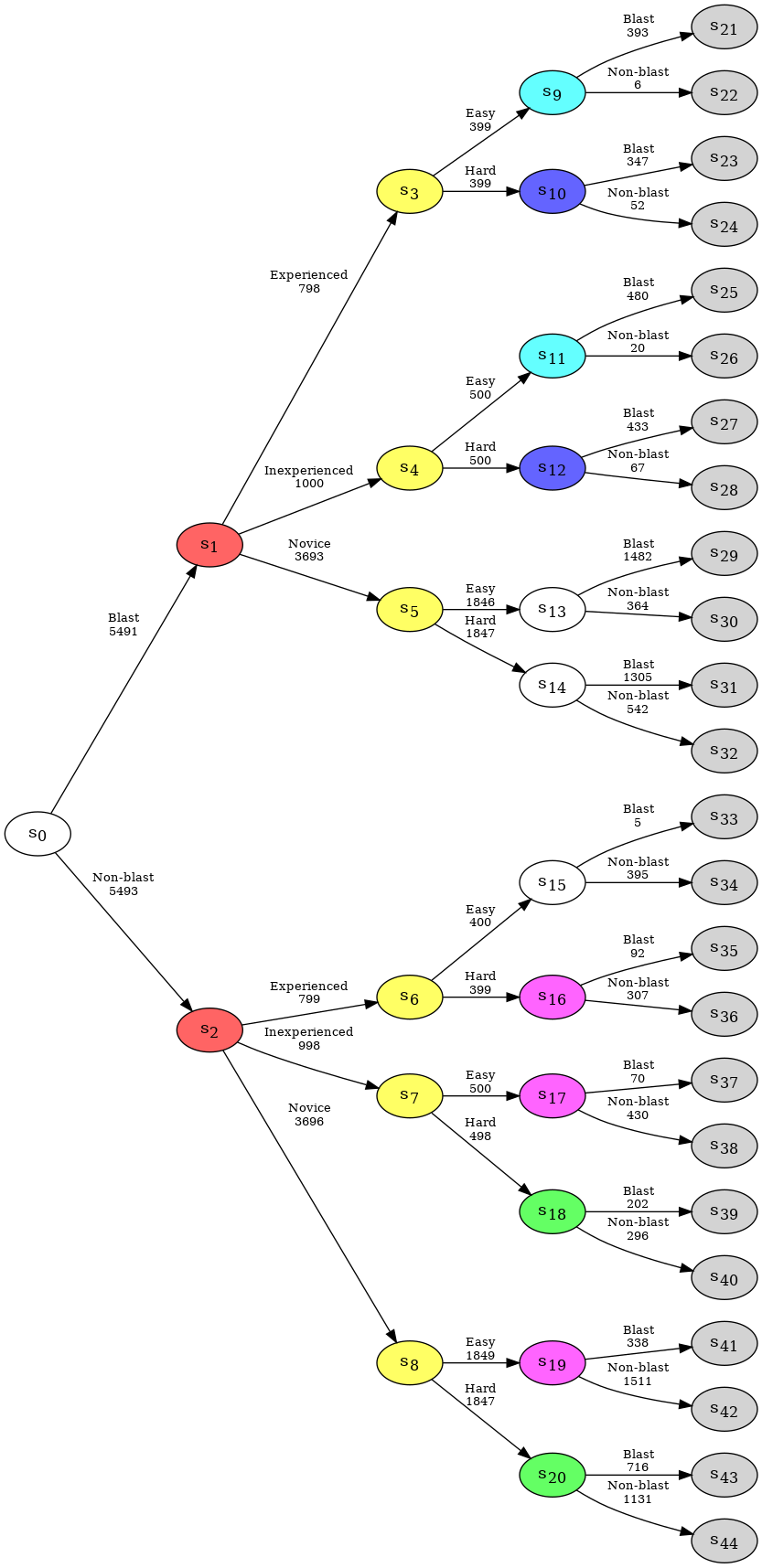
In an event tree, each situation is associated with a transition parameter vector which indicates the conditional probability of an individual, who has arrived at the situation, going along one of its edges. In order to create a CEG, we first need to elicit a staged tree. This is done by first partitioning situations into stages, which are collections of situations in the event tree whose immediate evolutions, i.e. their associated conditional transition parameter vectors, are equivalent. To indicate this symmetry, all situations in the same stage are assigned a single colour.
Identification of the stages in the event tree can be done using any suitable model selection algorithm. Currently, the only available selection algorithm in cegpy is the Agglomerative Hierarchical Clustering (AHC) algorithm (Freeman and Smith, 2011).
In order to create a staged tree in cegpy we first initialize a StagedTree object from the dataset and then run the AHC algorithm using the create_AHC_transitions method, as displayed below. The output of the AHC algorithm is a dictionary containing the following information:
Merged Situations- a list of tuples representing the partition of the nodes into stagesLog Likelihood- the log likelihood of the data under the model selected by AHC
from cegpy import StagedTree
st = StagedTree(df)
st.calculate_AHC_transitions()
{'Merged Situations': [('s1', 's2'),
('s20', 's18'),
('s10', 's12'),
('s7', 's4', 's3', 's5', 's8', 's6'),
('s9', 's11'),
('s17', 's19', 's16'),
('s0',),
('s13',),
('s14',),
('s15',)],
'Log Likelihood': -30091.353114865367}
Within cegpy, singleton stages, i.e. stages containing a single situation, are coloured white, leaves and their corresponding sink node are coloured in light-grey. Running AHC on our data set results in the following staged tree.
st.create_figure()
Custom Hyperstages#
cegpy allows the user to specify which situations are allowed to be merged by the AHC algorithm. This is done by specifying a hyperstage (Collazo et al., 2017) which is a collection of sets such that two situations cannot be in the same stage unless they belong to the same set in the hyperstage. Under a default setting in cegpy, all situations which have the same number of outgoing edges and equivalent set of edge labels are in the same set within the hyperstage. The default hyperstages of a given tree can be displayed by accessing the hyperstage property, which returns a list of lists, where each sublist contains situations belonging to the same hyperstage.
st.hyperstage
[['s0',
's9',
's10',
's11',
's12',
's13',
's14',
's15',
's16',
's17',
's18',
's19',
's20'],
['s1', 's2'],
['s3', 's4', 's5', 's6', 's7', 's8']]
In this example, situations \(s_1\) and \(s_2\) belong to the same hyperstage. Each of them has three emanating edges with labels Experienced, Inexperienced, and Novice. However, stages \(s_6\) and \(s_15\) belong to different hyperstages. They both have two emanating edges, yet different labels: Easy, Hard and Blast, Non-blast.
We can specify a different hyperstage at the point of running the AHC algorithm by passing a list defining the hyperstage partition as a parameter to the calculate_AHC_transitions method, for example:
new_hyperstage = [
['s0'],
['s3', 's4', 's5', 's6', 's7', 's8', 's9', 's10', 's11', 's12',
's13', 's14', 's15', 's16', 's17', 's18', 's19','s20'],
['s1', 's2'],
]
st.calculate_AHC_transitions(hyperstage=new_hyperstage)
{'Merged Situations': [('s1', 's2'),
('s20', 's18'),
('s10', 's12'),
('s7', 's4', 's3', 's5', 's8', 's6'),
('s9', 's11'),
('s17', 's19', 's16'),
('s0',),
('s13',),
('s14',),
('s15',)],
'Log Likelihood': -30091.353114865367}
Structural and sampling zeros / missing values#
The package, by default, treats all blank and NaN cells as structural missing values, i.e. data that is missing for a logical reason. However, sometimes these might occur due to sampling limitations; sampling missing values. We may also not observe a certain value for a variable in our data set (given its ancestral variables) not because that value is a structural zero but because of sampling limitations, in which case we are dealing with sampling zeros.
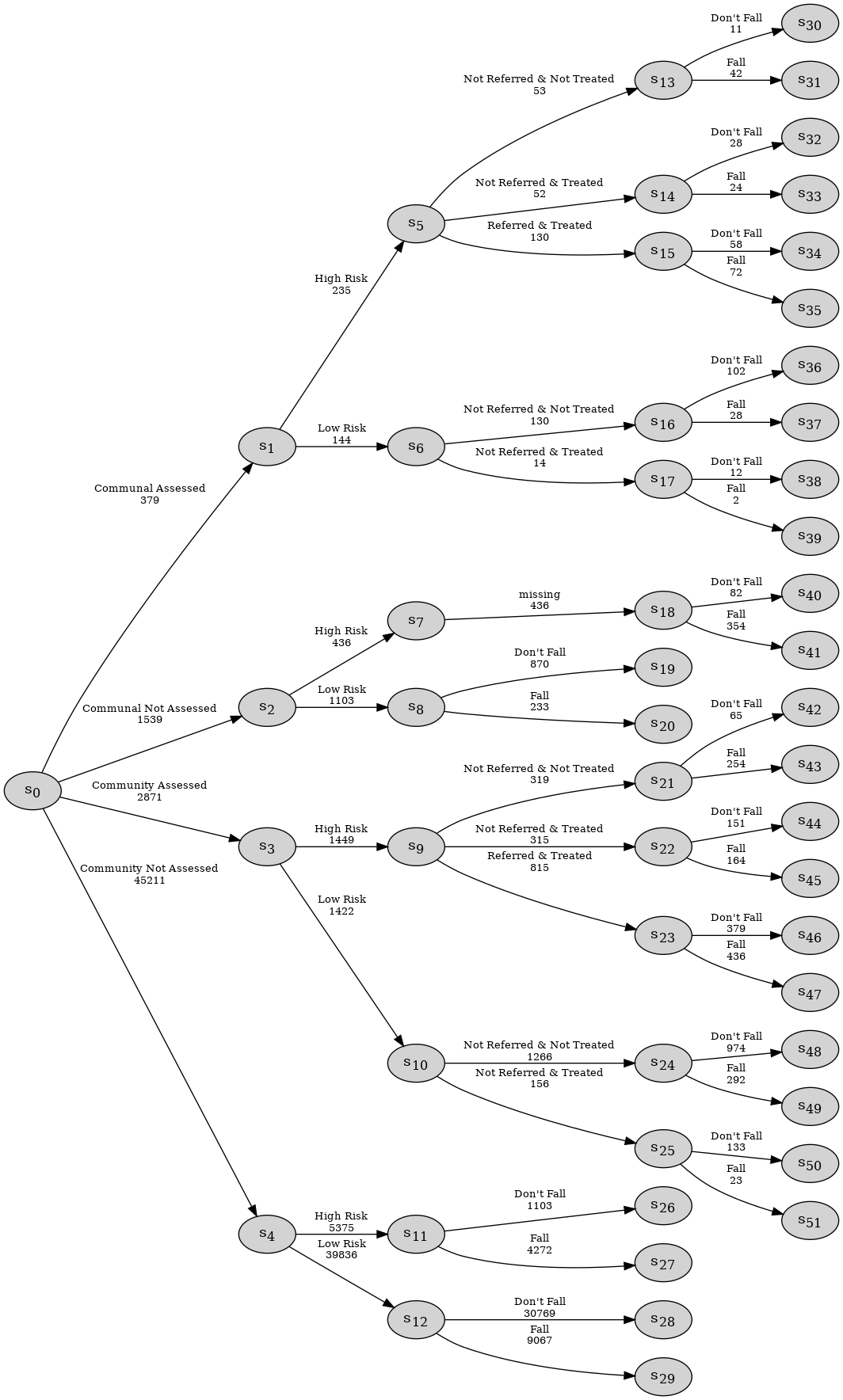
Consider the following example of the falls.xlsx data set which provides information concerning adults over the age of 65, and includes four categorical variables as given below with their state spaces:
Housing Assessment: Living situation and whether they have been assessed, state space:
{"Communal Assessed", "Communal Not Assessed", "Community Assessed", "Community Not Assessed"};Risk: Risk of a future fall, state space:
{"High Risk", "Low Risk"};Treatment: Referral and treatment status, state space:
{"Not Referred & Not Treated", "Not Referred & Treated", "Referred & Treated"};Fall: the outcome, state space:
{"Fall", "Don’t Fall"}.
df = pd.read_excel('../../data/Falls_Data.xlsx')
df.head()
| HousingAssessment | Risk | Treatment | Fall | |
|---|---|---|---|---|
| 0 | Community Not Assessed | Low Risk | NaN | Fall |
| 1 | Community Not Assessed | High Risk | NaN | Fall |
| 2 | Community Not Assessed | Low Risk | NaN | Don't Fall |
| 3 | Community Not Assessed | Low Risk | NaN | Don't Fall |
| 4 | Community Not Assessed | Low Risk | NaN | Fall |
et = EventTree(df)
et.create_figure()
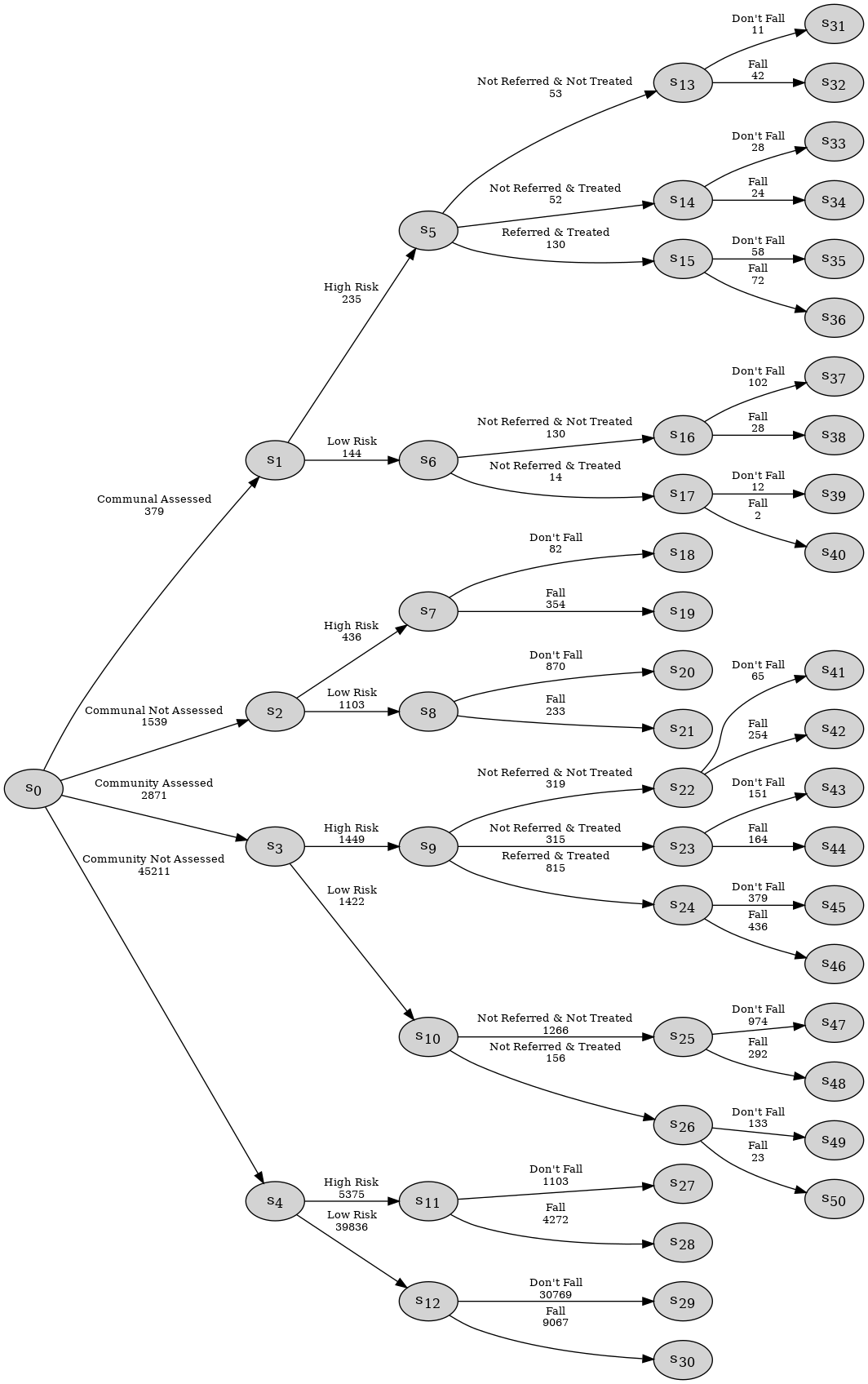
Observe that this process has structural asymmetries. None of the individuals assessed to be low risk are referred to the falls clinic and thus, for this group, the count associated with the _Referred & Treated’}$ category is a structural zero:
df[df.Risk == "Low Risk"]['Treatment'].value_counts()
Treatment
Not Referred & Not Treated 1396
Not Referred & Treated 170
Name: count, dtype: int64
Moreover, for individuals who are not assessed, their responses are structurally missing for the Treatment variable:
# Missing values in each column
print(df.isna().sum())
# Missing values for Treatment are structural,
# they are missing due to the lack of assessment:
df[df.HousingAssessment.isin([
'Community Not Assessed', 'Communal Not Assessed'
])]['Treatment'].isna().sum()
HousingAssessment 0
Risk 0
Treatment 46750
Fall 0
dtype: int64
46750
In cegpy any paths that should logically be in the event tree description of the process but are absent from the dataset due to sampling limitations would need to be manually added by the user using the sampling zero paths argument when initialising the EventTree object. Further, not all missing values in the dataset will be structurally missing.
How to distinguish between structural and sampling missing values?#
e.g. Falls example: Suppose that some individuals in communal establishments who are not formally assessed but are known to be high risk were actually either "Not Referred & Treated" or "Not Referred & Not Treated" but that these observations were missing in the falls.xlsx dataset due to sampling limitations. All the other blank/NaN cells are structurally missing.
idx = (df.HousingAssessment == 'Communal Not Assessed') & (df.Risk == 'High Risk')
df[idx]
| HousingAssessment | Risk | Treatment | Fall | |
|---|---|---|---|---|
| 67 | Communal Not Assessed | High Risk | NaN | Fall |
| 72 | Communal Not Assessed | High Risk | NaN | Fall |
| 95 | Communal Not Assessed | High Risk | NaN | Fall |
| 102 | Communal Not Assessed | High Risk | NaN | Fall |
| 132 | Communal Not Assessed | High Risk | NaN | Fall |
| ... | ... | ... | ... | ... |
| 49065 | Communal Not Assessed | High Risk | NaN | Fall |
| 49087 | Communal Not Assessed | High Risk | NaN | Fall |
| 49135 | Communal Not Assessed | High Risk | NaN | Don't Fall |
| 49461 | Communal Not Assessed | High Risk | NaN | Fall |
| 49905 | Communal Not Assessed | High Risk | NaN | Fall |
436 rows × 4 columns
To demarcate the difference between structural and sampling missing values, a user can give different labels to the structural and sampling missing values in the dataset and provide these labels to the struct_missing_label and missing_label arguments respectively when initialising the EventTree or StagedTree object.
In our example, we can replace the NaN values for the Treatment variable among the considered subset of data with a new label, e.g. samp_miss:
df.loc[idx, 'Treatment'] = 'samp_miss'
Next step is to tell the EventTree or StagedTree object about these missing value arguments as shown below. This will generate a new path along Communal Not Assessed', High Risk’, `missing’)}$:
et2 = EventTree(df,
missing_label='samp_miss',
)
et2.create_figure()
How to add sampling zeros?#
e.g. Falls example: Suppose that some individuals in the community who were assessed and high risk were referred and not treated. Suppose that our observations are still the same as in the falls.xlsx dataset. Here, by design, this was allowed, but was not observed in the dataset. So we need to add this value in manually as a path ("Community Assessed", "High Risk", "Referred & Not Treated"). We also need to add in the values that follow it: i.e. ("Community Assessed", "High Risk", "Referred & Not Treated", "Fall") and ("Community Assessed", "High Risk", "Referred & Not Treated", "Don't Fall").
In cegpy any paths that should logically be in the event tree description of the process but are absent from the dataset due to sampling limitations would need to be manually added by the user using the sampling zero paths argument when initialising the EventTree or StagedTree object. No changes need to be made to the dataset, as shown below:
st2 = StagedTree(df,
sampling_zero_paths=[
('Community Assessed', 'High Risk', 'Referred & Not Treated'),
('Community Assessed', 'High Risk', 'Referred & Not Treated', 'Fall'),
('Community Assessed', 'High Risk', 'Referred & Not Treated', "Don't Fall")
])
st2.calculate_AHC_transitions()
st2.create_figure()